We Can't Define a “Healthy Microbiome”
The composition of human gut microbial communities varies widely, but functional capacity is conserved
Universal interest in human-associated microbial communities in recent decades has galvanized both citizens and researchers alike to probe, analyze, and perhaps even biohack our bugs in an effort to potentially improve quality of life and ameliorate disease. Many commercial companies offer a glimpse into the inner workings of your microbial inhabitants through the use of at-home testing kits.
Countless others present opportunities to shift the microbial balance in one’s favor through the use of probiotics, prebiotics, synbiotics, phages, helminths, quorum sensing inhibitors, immune modulators, skin creams, body washes, vaginal suppositories, and more. However, our enthusiasm to learn and tweak our microbial communities should be tempered with caution and humility regarding how much remains unknown.
The compositions of gut microbial communities are highly variable across populations
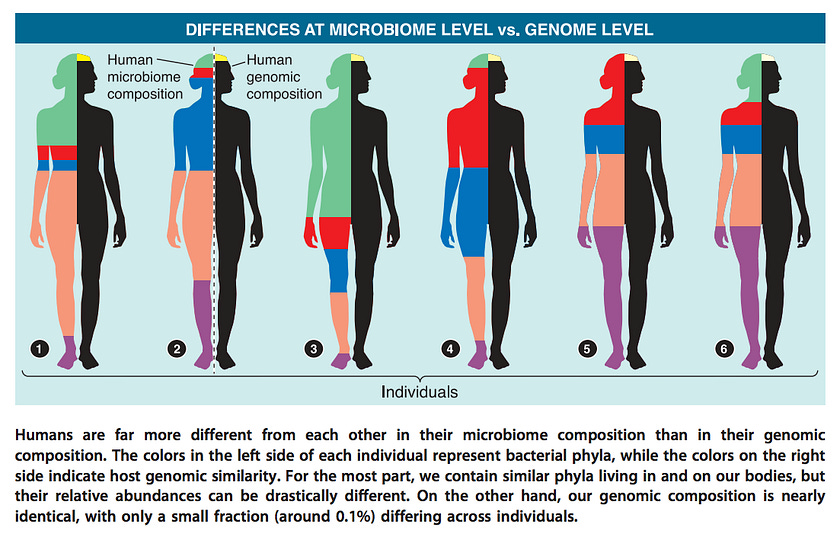
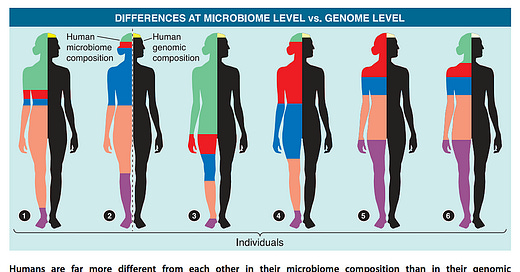
When it comes to composition, there is no singular healthy reference human microbiome. Which bacteria live in your gut depends on host genetics, ancestry, sex, diet, geographical considerations (sunlight exposure, climate), and many other factors. On average, any given two people share 99.9% of their genome but only about one third of their gut microbiome (by composition).
The latter finding generally holds true even for identical twins (37% vs 35% for unrelated individuals). The remaining two-thirds may be influenced by mode of birth delivery, breastfeeding history, age, stress, medication use, variability in sleep patterns, circadian rhythms, exercise, and other environmental factors.
Before the advent of large-scale microbiome sequencing efforts, researchers assumed the existence of a common “core” of microbes shared among human populations, as culture-based studies seemed to suggest that most adults harbored certain bacterial species such as Escherichia coli.
This expectation of a core microbiota was swiftly upended by the results of non-culture sequencing studies, which repeatedly demonstrated vast microbial diversity that is both highly variable over time and across human populations!
Moreover, since most microbiome studies disproportionately sample from WEIRD (Western, Education, Industrialized, Rich, Democratic) populations, we may still only be seeing the tip of the iceberg in terms of extant global microbial diversity.
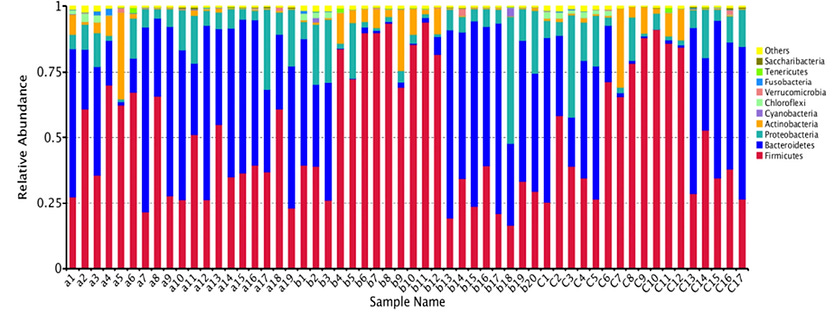
Even though two individuals may not share any of the same gut bacterial species, the types of bacteria able to successfully colonize the human gut belong to just a handful of taxonomic groups called phyla.
In this manner, the gut microbiota display phylogenetic underdispersion. In other words, out of the hundreds of bacterial phyla found in natural environments, the human gut is characterized by only 5–7 distinct phyla. However, the relative proportions of these phyla can vary considerably among individuals.
The composition of microbial communities may change, but function remains the same
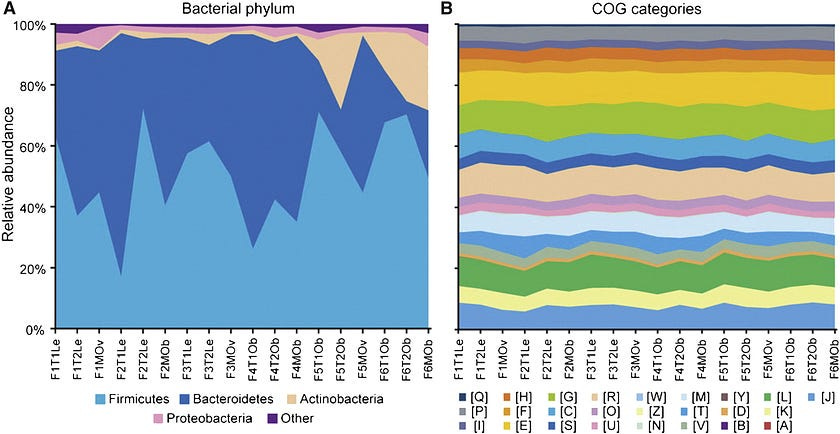
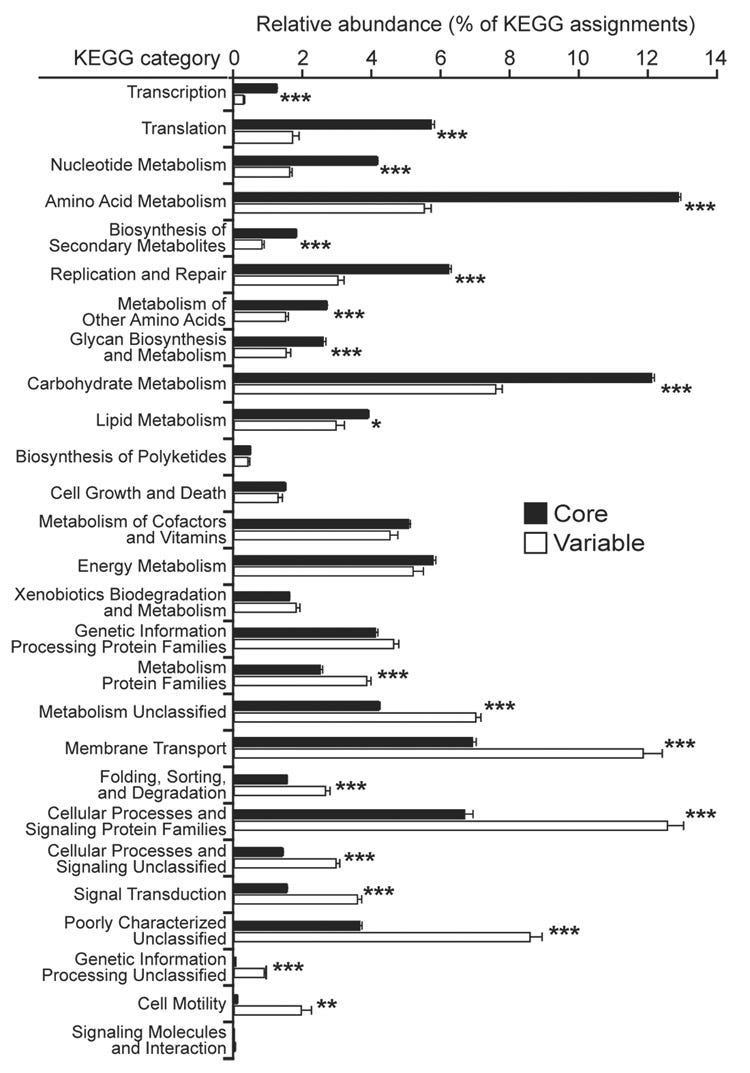
Although individuals exhibit a great deal of microbial variation at the compositional level, the functional profile of gut microbial communities remains fairly stable. In other words, while we may harbor markedly different microbes, the activities those microbes carry out are remarkably similar.
A 2012 study conducted by the Human Microbiome Project Consortium reported that genes representing metabolic pathways were stable among individuals despite variation in community structure. The Turnbaugh lab similarly described how microbial genes shared among individuals comprise “an extensive, identifiable ‘core microbiome’ at the gene, rather than at the organismal lineage, level.”
Overall, the metabolic capabilities of gut microbiomes seem relatively conserved among healthy adult individuals, and deviations from this core microbiome at the gene level are associated with obesity and other disease conditions. As Dr. Rob Knight of the American Gut Project summarizes,
“The common core is at the level of functions, not members.”
Tied to the overall conservation of functional genes is the concept of functional redundancy, a backup system in which many types of bacteria are capable of carrying out the same metabolic processes. As a result, a healthy gut microbiome can be assembled in a number of different ways.
For example, Oxalobacter formigenes is the most well-characterized oxalate-degrading microbe in the colon, and its presence has been associated with a reduced risk of developing kidney stones.
But certain strains of Lactobacillus and Bifidobacterium have also been shown to carry the same oxc and frc genes responsible for oxalate degradation as O. formigenes, and Eubacterium lentum and Enterococcus faecalis can consume oxalate as well.
Similarly, Roseburia intestinalis, Faecalibacterium prausnitzii, Butyricicoccus pullicaecorum, Eubacterium hallii, Anaerostipes caccae, and several other species are capable of synthesizing butyrate, a short-chain fatty acid.
This built-in redundancy ensures that the loss of one particular strain or species doesn’t spell metabolic doom for the host. Accordingly, testing services reporting that certain bacterial species are outside a specified abundance range should be interpreted with caution.
Functional redundancy confers stability and negates the need for keystone species, defined as those species that have a disproportionately large effect on community structure relative to their abundance.
Potential keystone species such as Akkermansia muciniphila, Faecalibacterium prausnitzii, Roseburia intestinalis, Christensenella minuta, and Bacteroides uniformis have been proposed, yet none can be generalized as fundamental across all human gut populations. The Hadza hunter-gatherers of Tanzania, for instance, do not harbor any Bifidobacterium but still have very low rates of chronic disease.
Variations in gut metabolism outside the core microbiome can result in individualized responses to food and drugs
While essential pathways, such as those responsible for the breakdown of carbohydrates and protein, may be conserved in the common core metagenome, or collective genetic material present among the microbiota, not all metabolic pathways are represented in the core. Differences in these variable pathways can affect individual responses to diet, medication, and other therapeutic interventions.
For example, the health benefits conferred by a soy-rich diet are mediated by the production of a bacterial metabolite known as S-(-)equol. However, as few as 25% of adults in North America and Europe produce this compound in response to dietary soy intake compared to as many as 80% of individuals in Japan, Korea, and China.
Thus, the cancer-protective effects of soy-based diets found in Asian populations may not translate to Western populations due to differences in gut microbial metabolic capacities. Similarly, the gut microbiota can affect how drugs such as acetaminophen, the active ingredient in Tylenol, are metabolized by the liver.
This knowledge creates opportunities for personalized interventions based on assessments of microbiome activity to maximize efficacy and minimize toxicity.
While the types of bacteria in people's guts can be very different, the functions they perform are remarkably similar, forming a core set of essential metabolic activities.
Integrative models that combine both compositional (“who they are”) and functional information (“what they do”) can inform more personalized therapeutic interventions going forward.




Great article, of course. It took me about two years of veggies and greens to get my microbiome to deal with the fiber in a whole foods plant based diet. I refuse to take any antibiotics for that reason, unless they are absolutely necessary, and that hasn't happened in many years. Thanks for another interesting, informative posting, Nita.
The body is such a mystery. Why are your mast cells not behaving themselves? Is there an answer to why one person is spared illness and not another? More philosophy than medicine.